Download the dataset
You can explore the dataset interactively using the neuPrint platform. To access the data programmatically, we recommend using the dedicated Python and R packages:
The neuprint-python package provides a Python
interface to the neuPrint API.
pip install neuprint-python
Next, go to neuPrint and create an account. Follow these instructions for getting your API token.
from neuprint import Client
client = Client("https://neuprint.janelia.org", api_token="<your_token>")
# Get neuron annotations and neuropil innervation
from neuprint import fetch_neurons
neurons, syndist = fetch_neurons("DNge104")
# Get connectivity
from neuprint import fetch_adjacencies
outgoing_edges, neuron_info = fetch_adjacencies("DNge104")
incoming_edges, neuron_info2 = fetch_adjacencies(None, "DNge104")
For more examples, please see the neuPrint Github repository.
If you want to work with neuron morphology, consider using navis. It wraps
the neuprint-python interface and adds functions to read skeletons and meshes as navis objects which
you can then use for visualization and analysis:
pip install navis[all]
>>> import navis
>>> import navis.interfaces.neuprint as neu
>>> client = Client("https://neuprint.janelia.org", api_token="<your_token>")
>>> skels = neu.fetch_skeletons(neu.NeuronCriteria(type="DNge104"))
>>> skels
<class 'navis.core.neuronlist.NeuronList'> containing 2 neurons (779.2KiB)
type name id ... cable_length soma units
0 navis.TreeNeuron DNge104_R 12781 ... 1558673.00 10 8 nanometer
1 navis.TreeNeuron DNge104_L 556329 ... 1690152.25 3 8 nanometer
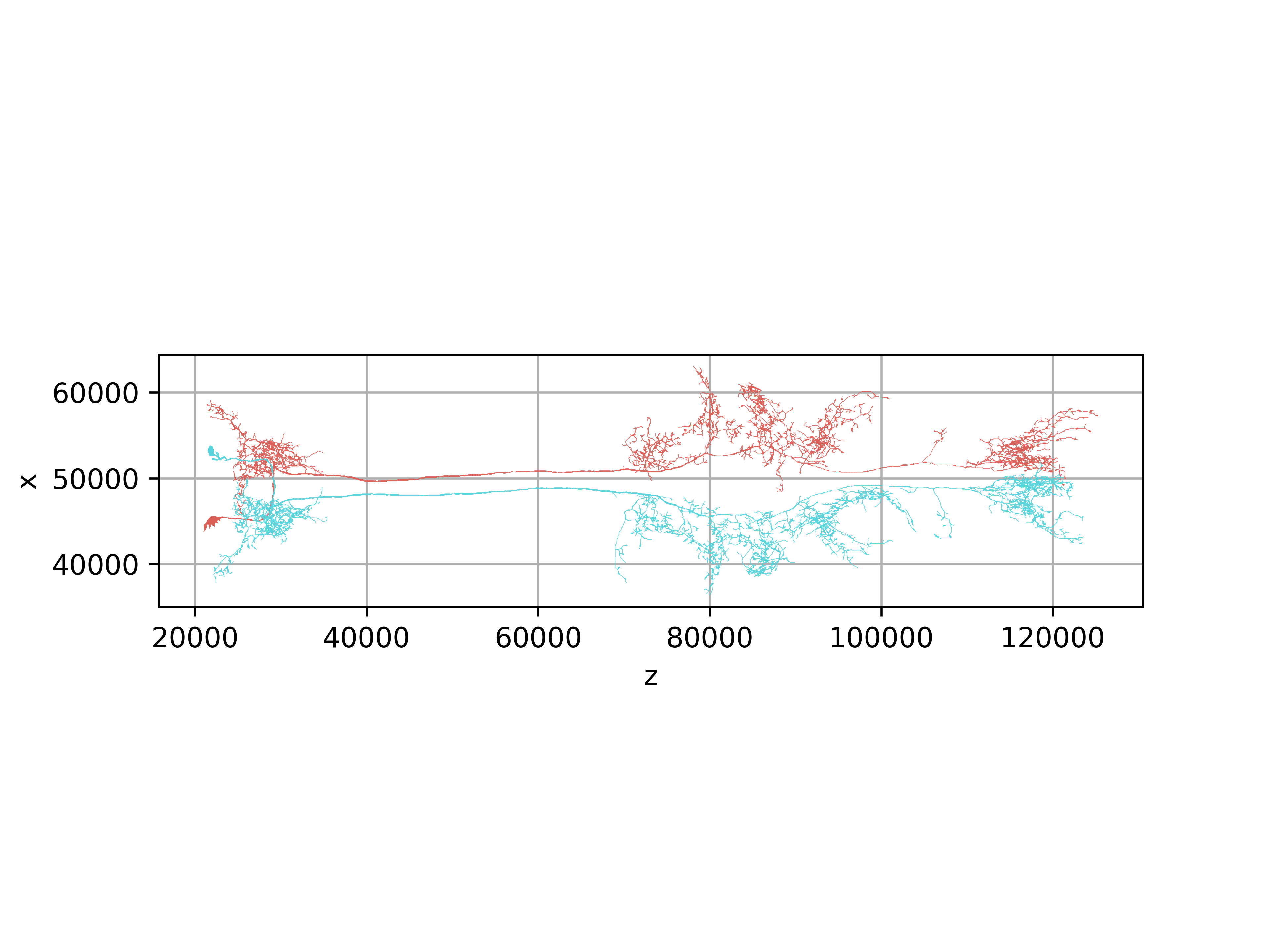
>>> fig, ax = navis.plot2d(skels, view=('z', 'x'), radius=True)
Please see the navis neuPrint tutorial
for more examples. Also check out the flybrains extension package and the corresponding
tutorial for transforming spatial data
(such as skeletons or meshes) between male CNS space and other common Drosophila template spaces.
The neuprintr package provides a programmatic
interface to the neuPrint API for R.
# install
if (!require("devtools")) install.packages("devtools")
devtools::install_github("natverse/neuprintr")
# use
library(neuprintr)
Next, go to neuPrint and create an account. Follow these instructions for getting your API token.
# Manually initialize the connection - alternatively you can set variables in your .Renviron file
conn = neuprint_login(server= "https://neuprint.janelia.org/", token= "<your_token>")
For more examples, please see the neuprintr documentation.
All connectome data is available for bulk download, suitable for analyses or ingestion into alternative databases.
Selected image and segmentation volumes are described below. Please inspect the data source URLs in the MaleCNS neuroglancer scene for related volumes.
Except where noted, the data is available in neuroglancer precomputed format, readable with tensorstore or cloud-volume. Links beginning with gs:// refer to Google Storage Bucket locations.
gs://flyem_cns_z0720_07m_dvidcoords_n5- The aligned EM volume, uncompressed and without CLAHE normalization, in N5 format (supported by tensorstore).
- 8nm isotropic resolution.
- Data type
uint8 - Neuroglancer view
gs://flyem-male-cns/em/em-clahe-jpeg- The aligned EM volume, contrast-adjusted with CLAHE normalization and JPEG-encoded.
- Ideal for interactive browsing.
- Data type
uint8
gs://flyem-male-cns/v0.9/segmentation- The
v0.9proofread neuron segmentation. - 8nm isotropic resolution.
- Voxels are stored as
uint64, but the upper 32 bits are never used.
- The
gs://flyem-male-cns/v0.9/malecns-v0.9-nuclei-seg-16nm- The
v0.9nuclear segmentation. Not filtered to exclude glia, noise, etc. These served as the basis for our cell body annotations, which were manually reviewed. - 16nm isotropic resolution.
- Voxel datatype is
uint64.
- The
gs://flyem-male-cns/rois/fullbrain-roi-v4- Brain neuropil compartment segmentation, initialized via transfer from ROIs in JRC2018M and refined manually.
- 256nm isotropic resolution
- Voxels are stored as
uint64, but max value is96.
gs://flyem-male-cns/rois/malecns-vnc-neuropil-roi-v0- VNC neuropil compartment segmentation, refined manually.
- 256nm isotropic resolution
- Voxels are stored as
uint64, but max value is27.
Example code for reading the EM image data using Python and cloud-volume:
>>> from cloudvolume import CloudVolume
>>> # Initialize a CloudVolume for the EM image data
>>> vol = CloudVolume('precomputed://gs://flyem-male-cns/em/em-clahe-jpeg', use_https=True)
>>> vol.shape
(94088, 78317, 134576, 1)
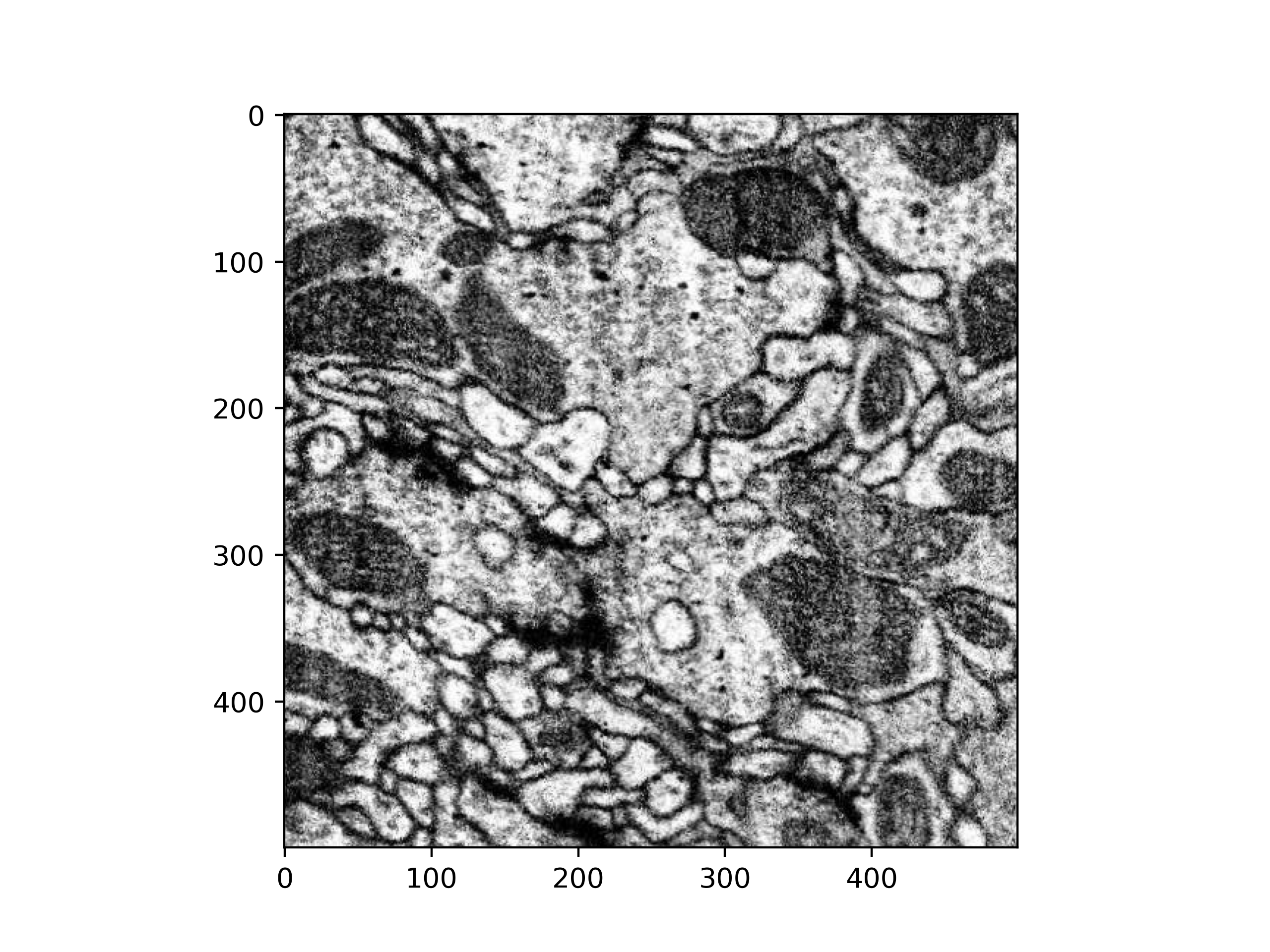
>>> # Grab a 500x500 voxels cutout
>>> cutout = vol[40000:40500, 40000:40500, 20000, 0]
>>> # Plot a single slice using matplotlib
>>> import matplotlib.pyplot as plt
>>> plt.imshow(cutout[:, :, 0, 0], cmap='gray')
Neuron-level annotations are provided in Apache Arrow Feather file format, which can be read
using e.g. pyarrow (Python), pandas (Python) or the arrow package (R).
- body-annotations-male-cns-v0.9-minconf-0.5.feather
- Curated neuron annotations (classes, types, sides, etc.), excluding neurotransmitter properties.
- 13 MB
- body-neurotransmitters-male-cns-v0.9.feather
- Aggregate neurotransmitter predictions for each neuron. See manuscript methods section for details.
- 42 MB
- body-stats-male-cns-v0.9-minconf-0.5.feather
- summary statistics (synapse counts) of all segments in the dataset (excluding those with no synapses)
- 780 MB
For updated annotations for the female brain "FlyWire" connectome, please see the flyconnectome/flywire_annotations Github repository.
We provide several tables describing synaptic connectivity in the MaleCNS dataset:
- connectome-weights-male-cns-v0.9-minconf-0.5.feather
- segment-to-segment connection strengths for all segments in the dataset (excluding those with no synapses)
- This is the full connection graph.
- 1.1 GB
- syn-points-male-cns-v0.9-minconf-0.5.feather
- Pre-synapse and post-synapse locations, body (segment) ID, and encompassing ROIs
- Pre-synapses and post-synapses are listed in separate rows. The 'kind' column indicates whether the point is
PreSynorPostSyn. - Pre-synapses and post-synapses are uniquely identified by their
x,y,zlocations (expressed in voxel units, i.e. 8nm). - In the partner table (below), a single pre-synapse may connect to multiple post-synapses. In this table, each unique pre-synaptic location is listed only once.
- 12.7 GB
- syn-partners-male-cns-v0.9-minconf-0.5.feather
- Synaptic partner pairs, along with their associated body IDs and primary neuropil.
- Columns:
'x_pre', 'y_pre', 'z_pre', 'body_pre', 'conf_pre', 'x_post', 'y_post', 'z_post', 'body_post', 'conf_post', 'primary_post' - 6.8 GB
- tbar-neurotransmitters-male-cns-v0.9.feather
- Neurotransmitter prediction probabilities for each pre-synapse
- 2.7 GB
Centerline skeletons for all neurons can be downloaded in several formats and coordinate spaces:
gs://flyem-male-cns/v0.9/segmentation/skeletons-malecns/skeletons-swc/- Directory of neuron skeletons in SWC format, with names such as
12781.swc. - Male CNS EM coordinate space, with coordinates specified in 8nm units.
- Directory of neuron skeletons in SWC format, with names such as
gs://flyem-male-cns/v0.9/segmentation/skeletons-malecns/skeletons-precomputed/- Same as above, but in neuroglancer's
precomputedskeleton format (unsharded). - Male CNS EM coordinate space, coordinates in 1 nm units.
- Same as above, but in neuroglancer's
gs://flyem-male-cns/v0.9/segmentation/skeletons-malecns-mirrored/skeletons-swc/- Directory of mirrored neuron skeletons in SWC format, with names such as
12781.swc. - Mirrored using a transform available via
navis-flybrains. - 8nm units
- Directory of mirrored neuron skeletons in SWC format, with names such as
gs://flyem-male-cns/v0.9/segmentation/skeletons-malecns-mirrored/skeletons-precomputed/- Same as above, but in neuroglancer's
precomputedskeleton format (unsharded). - 1nm units
- Same as above, but in neuroglancer's
gs://flyem-male-cns/v0.9/segmentation/skeletons-unisex-template/- Directory of MaleCNS skeletons transformed to JRC2018 unisex template space.
- Transform available via
navis-flybrains. - 1um (micron) units
The Male CNS is licensed under CC-BY.
If you want to setup your own neuprint instance or explore the data using neo4j directly, you can download the neo4j database and input CSV files used to construct it:
gs://flyem-male-cns/v0.9/database/neo4j- The complete neo4j database backing the
male-cns:v0.9neuprint dataset hosted on neuprint.janelia.org - Built for neo4j v4.4.16
- Constructed using
flyem-snapshot
- The complete neo4j database backing the
gs://flyem-male-cns/v0.9/database/neuprint-inputs- The input CSV files used to construct the neo4j database.
This project is a collaboration between FlyEM (HHMI Janelia), the University of Cambridge (Dept. of Zoology), the MRC Laboratory of Molecular Biology, and Google Research.
The Male CNS dataset is licensed under CC-BY.